MDClone is a free, secure, self-service platform for building queries and downloading computationally derived (“synthetic”) data from the institute’s research data core (RDC). Since the data do not contain protected health information (PHI), their use is not classified as human subject research.
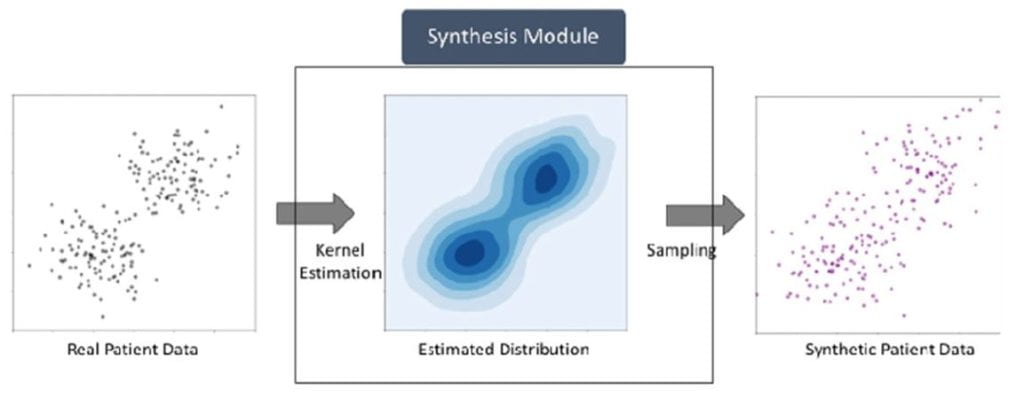
The Institute for Informatics, Data Science and Biostatistics (I2DB) implemented the MDClone platform in 2018, and conducted a landmark study validating that synthetic data produces the same results as original data while maintaining data privacy. The institute’s significant effort and investment in securing this resource for our campus is part of a strategic goal to accelerate the pace of data-driven research at Washington University in St. Louis.
Our instance of MDClone is designed to meet the specific clinical and translational research needs of researchers at the School of Medicine.
User support is provided by MDClone. Contact the Clinical Specialist from MDClone, Michelle Schneider at michelle.schneider@mdclone.com for questions regarding using MDClone
The I2DB scope of support includes:
- Online training and onboarding to the platform
- Demonstration videos
- Data brokerage services for downloading the original data (requires IRB approval)
MDClone data coverage includes:
- Inpatient and outpatient data from across BJC network facilities
- Demographics, encounters, diagnosis, procedure, medication order, lab results, vitals, allergies, surgery, microbiology, social history, problem list, practitioner, and insurance
- Data from 2010 to present
For a more detailed breakdown of the data available in MDClone, please review the MDClone Data Inventory.
Levels of MDClone Data Accessibility and Pricing
MDClone creates computationally derived or synthetic datasets from real patient data stored in a synthetic datalake. The datalake data consists of a limited EPIC dataset that is ingested monthly. This creates three different MDClone Levels of data available for users. A brief description of these levels and how to access them is below:
Synthetic datasets can be accessed for free by Washington University researchers with access to MDClone. There is no limit to the number of queries you can create and synthetic datasets you can download. Note that some large queries (i.e., More than 1 million cohort or including hundreds of columns in your dataset) may have issues generating.
- Obtain access to MDClone
- Build a query
- Generate and download synthetic data
Data ingested into the MDClone data lake is a limited EHR dataset. The data are considered limited because 1) all PHI is stripped, 2) MRN numbers are hashed, 3) dates are shifted, and 4) only structured data from the medical record (i.e., no notes, images, etc.) are ingested. Review of the IRB application and extraction of the limited data set are completed by the data brokerage team at a $150 per hour fee. Follow these steps to access a limited dataset:
- Complete steps 1-3 under synthetic data
- Obtain IRB approval
- Complete the I2DB Central Request Form, which will include IRB application number and status
- Receive a follow up email with an estimated cost or a request for additional details
- Send MDClone query to Data Brokerage
- Receive the dataset in a WUSTL Box folder
- For most queries, this will require 1-3 weeks to complete steps 4-7 and about $450
Following analyses of synthetic data, researchers may wish to obtain additional data for their cohort that are unavailable in the MDClone data lake. For example, if you create a query looking at total knee reconstruction, you may wish to obtain MRI imaging for patients in your cohort. It is not possible to ingest imaging data into MDClone; however, you can work with the data brokerage team to obtain this data for patients in your cohort. Review of IRB application and extraction of the limited data set is completed by the data brokerage team at a $150 per hour fee. Follow these steps to access additional, unstructured data for patients in your cohort
- Complete steps 1-3 under synthetic data
- Obtain IRB approval
- Complete the I2DB Central Request Form, which will include IRB application number and status
- Have a consultation with data brokerage
- Receive Statement of Work (SOW) with estimated total cost
- Total Hours X Estimated Hours of Effort = Total Cost
- Send MDClone query to Data Brokerage
- Receive the dataset in a WUSTL Box folder
- For most queries, this will require 7-10 weeks to complete steps 4-7 and unmask the limited dataset and $750-$1500
ICTS JIT funds can be used to cover the costs of the data pull. The 1-hour consultation, along with their Statement of Work (SOW), will help you to know what the budget will look like for the JIT.
I2DB publications related to MDClone:
- Understanding the opportunity and application of synthetic data in healthcare Paediatric and Perinatal Epidemiology
- Spot the Difference: Comparing Results of Analyses from Real Patient Data and Synthetic Derivatives. JAMIA Open
- The Use of Synthetic Electronic Health Record Data and Deep Learning to Improve Timing of High-Risk Heart Failure Surgical Intervention by Predicting Proximity to Catastrophic Decompensation. Frontiers in Digital Health
- Predicting Mortality among Patients with Liver Cirrhosis in Electronic Health Records with Machine Learning. PLoS One
- The National COVID Cohort Collaborative: Analyses of Original and Computationally Derived Electronic Health Record Data. Journal of Medical Internet Research
For more information, please contact us at i2help@wustl.edu.